Introduction and Orientation
M Hallett
21/07/2020
Outline for today
Introduce myself
Provide context for the course
Discuss logistics of the course
Discuss what is expected of you and I
Me
https://mikehallett.science My lab website
Data science for biologists Course Website
\({\tt michael.hallett@concordia.ca}\) e-mail
\({\tt @hallettmichael}\) Twitter

Early ’90s

Mid- to Late-’90s



Even for the bacteria like H. influenzae, slow-throughput technologies were simply not enough to sequence complete genomes. It took high-throughput approaches to make it feasible.
\(1,800,000 = 1.8 \cdot 10^6\) vs \(3,600,000,000 = 3.6 \cdot 10^9\) base pairs. Three orders of magnitude in \(5\) years.
Rapid growth and diversification
In addition to increased sequencing capacity, many other high-throughput technologies that measured different levels of the centra dogma of biochemistry came on-line.


And many additional “-omics” approaches.
Technology based on robotics, micromanipulation, nanofabrication, lasers, photolithography, microfluidics, microscopy …
Driven by a better understanding of protein and small molecule biochemistry, and a robust calculus of ways to manipulate nucleic acids (molecular biology).
2000’s
I moved to McGill as a professor in 2000. Became interested in cancer genomics and informatics.
The early 2000s saw a commoditization of equipment, the ability to apply these technologies in human contexts, and the development of so-called core facilities.

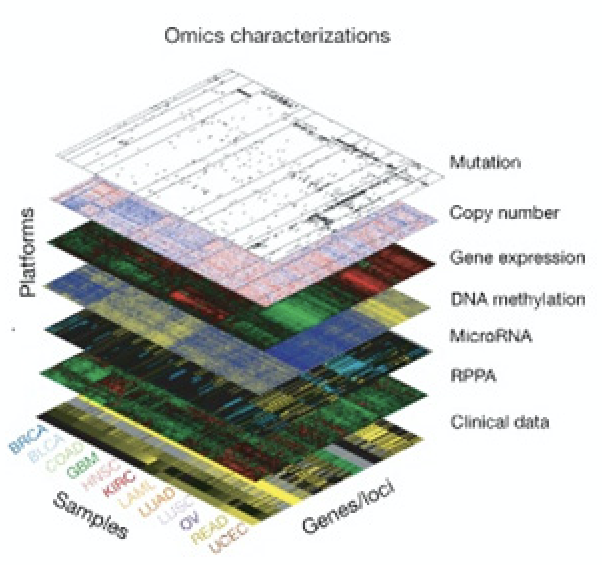
2010’s
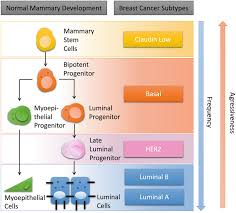
Moved to Concordia in 2018. We are interested in how cells are wired (the molecular network), how this wiring can change in response to stimuli, and how the changes in the network provide cells with advantages.


Bioinformatics, Computational Biology and Data Science
[Data Science in Biology] The ability to develop software and analytic approaches to wrangle with biological data, clean it up, kick it into shape and visualize it in a way that is both informative and honest.
[Bioinformatics] The development of tools, portals and databases to make biological information available to all life scientists.
[Computational Biology] The development of new analytic techniques, typically expressed as computer programs, to explore biological data and test hypotheses.
This course will you give you the fundamentals in all three disciplines with an emphasis on data science.
Computational Biologist
Ice Hockey \(\mapsto\) huh?
Hot Sun, Hard Rock… and many other useless adjectives.
Field Hockey \(\mapsto\) unfortunate misuse of word “hockey”.
Field Ball-Club Game \(\checkmark\)
(Road Hockey … ok English is weird sometimes … a valid Canadian game in summer)
\(\Rightarrow\) Hockey is ice hockey, therefore “ice” is obvious from the context and therefore no need to state explicitly.
IceHockeyComputational Biologist \(\mapsto\) huh?
Non-computational Biologist ?, Luddite Biologist ?
Cave Biolgist ?
\(\Rightarrow\) All biology is about information/computation, therefore all biologists are computational biologists (maybe just not good ones)
ComputationalBiologist
Much information “hidden” in bioinformatics databases
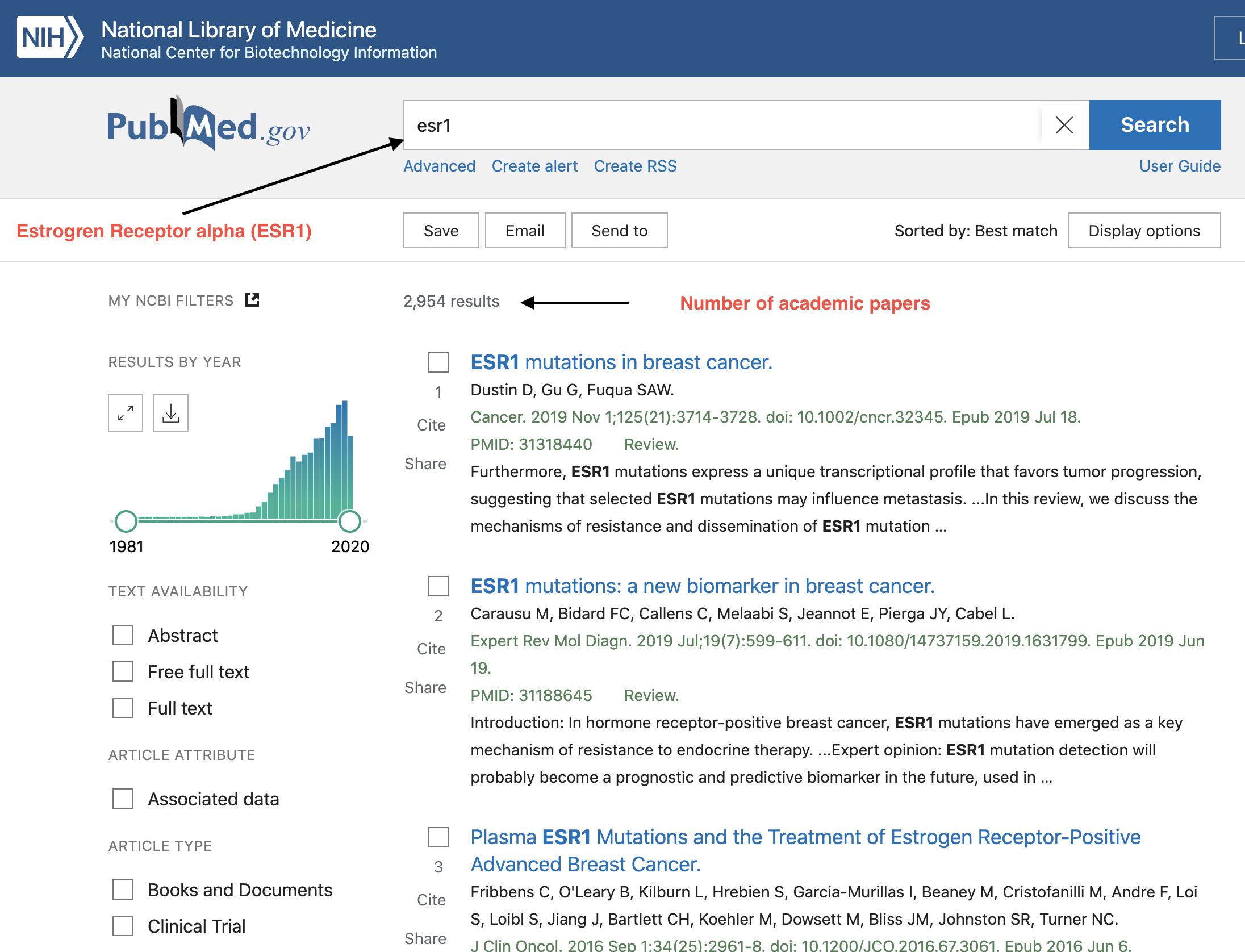
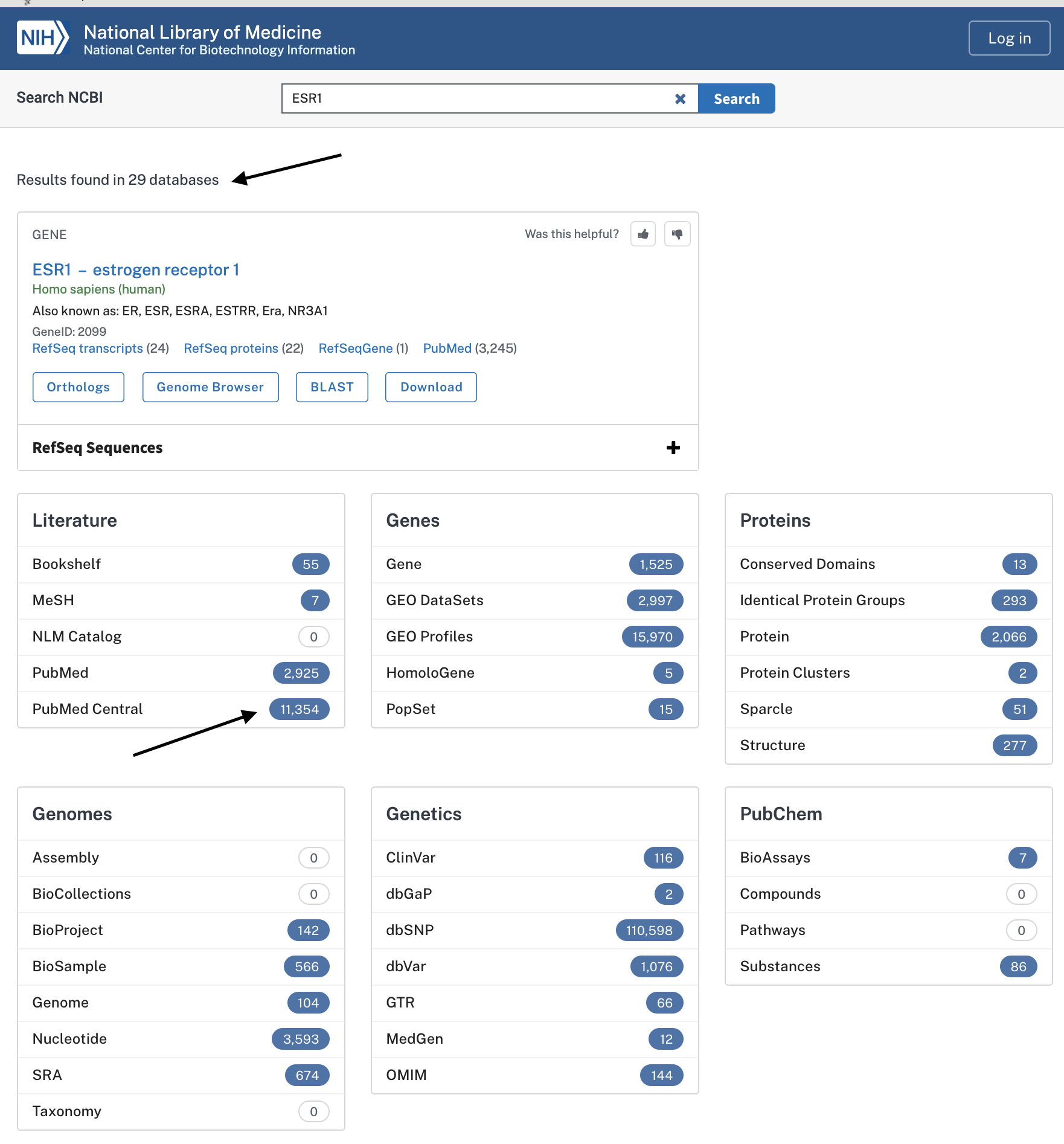
… everytime a human cell is sequenced ESR1 or its gene products are profiled.
Clearly, if you lack bioinformatic and data science skills, you are restricted to just reading manuscripts.
Outline for today
Introduce myself \(\checkmark\)
Provide context for the course \(\checkmark\)
Discuss logistics of the course \(\Leftarrow\)
Discuss what is expected of you and \(\Leftarrow\)
… but before re-directing to our course website, …
Some advice
- Bioinformatics, computational biology, data science are important.
- You need these skills; you cannot realistically expect to participate in modern biology without them.
- The good news is that data science is an experimental science, just like what you are used to.
- The primary means of doing experiments is through statistics and computation.
- This requires programming.
- Learning to program is like learning a natural language.
- It is a serious investment. It takes an open mind and practice.
BIOL480
© M Hallett, 2020 Concordia University
