Genome Annotations
M Hallett
21/07/2020
Biostrings: tools to manipule genetic strings
Last lecture we finished with a partial exploration of the Biostrings package in R.
We saw various functions applied to \({\tt DNAstring}\) datastructures.
… and we used the \({\tt BSgenome}\) package to quite seamlessly import full genomes into R. (Naming convention: \({\tt BSgenome.Organism.Provider.BuildVersion}\))
dnastring = DNAString("ATGATAAAAGAA")
dnastring
## 12-letter DNAString object
## seq: ATGATAAAAGAA
length(dnastring)
## [1] 12
reverseComplement(dnastring)
## 12-letter DNAString object
## seq: TTCTTTTATCAT
translate(dnastring)
## 4-letter AAString object
## seq: MIKE
library(BSgenome.Scerevisiae.UCSC.sacCer2)
yeast_1 <- Scerevisiae$chrI;
yeast_1
## 230208-letter DNAString object
## seq: CCACACCACACCCACACACCCACACACCACACCACA...GGTGTGTGGGTGTGGTGTGGGTGTGGTGTGTGTGGG
dinucleotideFrequency(yeast_1, as.prob=TRUE)
## AA AC AG AT CA CC CG
## 0.10400639 0.05426855 0.05916849 0.08587489 0.06614482 0.04005091 0.03078534
## CT GA GC GG GT TA TC
## 0.05695743 0.06287819 0.03871298 0.04099788 0.05620594 0.07028891 0.06090171
## TG TT
## 0.06784763 0.10490993Biostrings
String (Biostrings)
– Virtual class
– Concrete subclasses:
-- BString – Any “biological” sequence -- DNAString – DNA sequence -- RNAString – RNA sequence -- AAString – Amino acid sequence
The BSgenome series of packages
- BSgenome is a series of packages that import the complete genome of different organisms into R and represents them as Biostrings.
# BiocManager::install("BSgenome")
# BiocManager::install("BSgenome.Hsapiens.UCSC.hg19")
library(BSgenome)
available.genomes()
## [1] "BSgenome.Alyrata.JGI.v1"
## [2] "BSgenome.Amellifera.BeeBase.assembly4"
## [3] "BSgenome.Amellifera.UCSC.apiMel2"
## [4] "BSgenome.Amellifera.UCSC.apiMel2.masked"
## [5] "BSgenome.Aofficinalis.NCBI.V1"
## [6] "BSgenome.Athaliana.TAIR.04232008"
## [7] "BSgenome.Athaliana.TAIR.TAIR9"
## [8] "BSgenome.Btaurus.UCSC.bosTau3"
## [9] "BSgenome.Btaurus.UCSC.bosTau3.masked"
## [10] "BSgenome.Btaurus.UCSC.bosTau4"
## [11] "BSgenome.Btaurus.UCSC.bosTau4.masked"
## [12] "BSgenome.Btaurus.UCSC.bosTau6"
## [13] "BSgenome.Btaurus.UCSC.bosTau6.masked"
## [14] "BSgenome.Btaurus.UCSC.bosTau8"
## [15] "BSgenome.Btaurus.UCSC.bosTau9"
## [16] "BSgenome.Carietinum.NCBI.v1"
## [17] "BSgenome.Celegans.UCSC.ce10"
## [18] "BSgenome.Celegans.UCSC.ce11"
## [19] "BSgenome.Celegans.UCSC.ce2"
## [20] "BSgenome.Celegans.UCSC.ce6"
## [21] "BSgenome.Cfamiliaris.UCSC.canFam2"
## [22] "BSgenome.Cfamiliaris.UCSC.canFam2.masked"
## [23] "BSgenome.Cfamiliaris.UCSC.canFam3"
## [24] "BSgenome.Cfamiliaris.UCSC.canFam3.masked"
## [25] "BSgenome.Cjacchus.UCSC.calJac3"
## [26] "BSgenome.Dmelanogaster.UCSC.dm2"
## [27] "BSgenome.Dmelanogaster.UCSC.dm2.masked"
## [28] "BSgenome.Dmelanogaster.UCSC.dm3"
## [29] "BSgenome.Dmelanogaster.UCSC.dm3.masked"
## [30] "BSgenome.Dmelanogaster.UCSC.dm6"
## [31] "BSgenome.Drerio.UCSC.danRer10"
## [32] "BSgenome.Drerio.UCSC.danRer11"
## [33] "BSgenome.Drerio.UCSC.danRer5"
## [34] "BSgenome.Drerio.UCSC.danRer5.masked"
## [35] "BSgenome.Drerio.UCSC.danRer6"
## [36] "BSgenome.Drerio.UCSC.danRer6.masked"
## [37] "BSgenome.Drerio.UCSC.danRer7"
## [38] "BSgenome.Drerio.UCSC.danRer7.masked"
## [39] "BSgenome.Dvirilis.Ensembl.dvircaf1"
## [40] "BSgenome.Ecoli.NCBI.20080805"
## [41] "BSgenome.Gaculeatus.UCSC.gasAcu1"
## [42] "BSgenome.Gaculeatus.UCSC.gasAcu1.masked"
## [43] "BSgenome.Ggallus.UCSC.galGal3"
## [44] "BSgenome.Ggallus.UCSC.galGal3.masked"
## [45] "BSgenome.Ggallus.UCSC.galGal4"
## [46] "BSgenome.Ggallus.UCSC.galGal4.masked"
## [47] "BSgenome.Ggallus.UCSC.galGal5"
## [48] "BSgenome.Ggallus.UCSC.galGal6"
## [49] "BSgenome.Hsapiens.1000genomes.hs37d5"
## [50] "BSgenome.Hsapiens.NCBI.GRCh38"
## [51] "BSgenome.Hsapiens.UCSC.hg17"
## [52] "BSgenome.Hsapiens.UCSC.hg17.masked"
## [53] "BSgenome.Hsapiens.UCSC.hg18"
## [54] "BSgenome.Hsapiens.UCSC.hg18.masked"
## [55] "BSgenome.Hsapiens.UCSC.hg19"
## [56] "BSgenome.Hsapiens.UCSC.hg19.masked"
## [57] "BSgenome.Hsapiens.UCSC.hg38"
## [58] "BSgenome.Hsapiens.UCSC.hg38.masked"
## [59] "BSgenome.Mdomestica.UCSC.monDom5"
## [60] "BSgenome.Mfascicularis.NCBI.5.0"
## [61] "BSgenome.Mfuro.UCSC.musFur1"
## [62] "BSgenome.Mmulatta.UCSC.rheMac10"
## [63] "BSgenome.Mmulatta.UCSC.rheMac2"
## [64] "BSgenome.Mmulatta.UCSC.rheMac2.masked"
## [65] "BSgenome.Mmulatta.UCSC.rheMac3"
## [66] "BSgenome.Mmulatta.UCSC.rheMac3.masked"
## [67] "BSgenome.Mmulatta.UCSC.rheMac8"
## [68] "BSgenome.Mmusculus.UCSC.mm10"
## [69] "BSgenome.Mmusculus.UCSC.mm10.masked"
## [70] "BSgenome.Mmusculus.UCSC.mm8"
## [71] "BSgenome.Mmusculus.UCSC.mm8.masked"
## [72] "BSgenome.Mmusculus.UCSC.mm9"
## [73] "BSgenome.Mmusculus.UCSC.mm9.masked"
## [74] "BSgenome.Osativa.MSU.MSU7"
## [75] "BSgenome.Ptroglodytes.UCSC.panTro2"
## [76] "BSgenome.Ptroglodytes.UCSC.panTro2.masked"
## [77] "BSgenome.Ptroglodytes.UCSC.panTro3"
## [78] "BSgenome.Ptroglodytes.UCSC.panTro3.masked"
## [79] "BSgenome.Ptroglodytes.UCSC.panTro5"
## [80] "BSgenome.Ptroglodytes.UCSC.panTro6"
## [81] "BSgenome.Rnorvegicus.UCSC.rn4"
## [82] "BSgenome.Rnorvegicus.UCSC.rn4.masked"
## [83] "BSgenome.Rnorvegicus.UCSC.rn5"
## [84] "BSgenome.Rnorvegicus.UCSC.rn5.masked"
## [85] "BSgenome.Rnorvegicus.UCSC.rn6"
## [86] "BSgenome.Scerevisiae.UCSC.sacCer1"
## [87] "BSgenome.Scerevisiae.UCSC.sacCer2"
## [88] "BSgenome.Scerevisiae.UCSC.sacCer3"
## [89] "BSgenome.Sscrofa.UCSC.susScr11"
## [90] "BSgenome.Sscrofa.UCSC.susScr3"
## [91] "BSgenome.Sscrofa.UCSC.susScr3.masked"
## [92] "BSgenome.Tgondii.ToxoDB.7.0"
## [93] "BSgenome.Tguttata.UCSC.taeGut1"
## [94] "BSgenome.Tguttata.UCSC.taeGut1.masked"
## [95] "BSgenome.Tguttata.UCSC.taeGut2"
## [96] "BSgenome.Vvinifera.URGI.IGGP12Xv0"
## [97] "BSgenome.Vvinifera.URGI.IGGP12Xv2"
## [98] "BSgenome.Vvinifera.URGI.IGGP8X"
library(BSgenome.Hsapiens.UCSC.hg19)
print(Hsapiens)
## Human genome:
## # organism: Homo sapiens (Human)
## # provider: UCSC
## # provider version: hg19
## # release date: June 2013
## # release name: Genome Reference Consortium GRCh37.p13
## # 298 sequences:
## # chr1 chr2 chr3
## # chr4 chr5 chr6
## # chr7 chr8 chr9
## # chr10 chr11 chr12
## # chr13 chr14 chr15
## # ... ... ...
## # chr19_gl949749_alt chr19_gl949750_alt chr19_gl949751_alt
## # chr19_gl949752_alt chr19_gl949753_alt chr20_gl383577_alt
## # chr21_gl383578_alt chr21_gl383579_alt chr21_gl383580_alt
## # chr21_gl383581_alt chr22_gl383582_alt chr22_gl383583_alt
## # chr22_kb663609_alt
## # (use 'seqnames()' to see all the sequence names, use the '$' or '[[' operator
## # to access a given sequence)
print(Hsapiens$chr22)
## 51304566-letter DNAString object
## seq: NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN...NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGSgenome and Biostrings
- See if you can figure out the \({\tt sapply}\) function.
sapply(seqnames(Hsapiens),
function(seqname)
alphabetFrequency(Hsapiens[[seqname]], baseOnly=TRUE, as.prob = TRUE))
## chr1 chr2 chr3 chr4 chr5 chr6
## A 0.26307213 0.29236355 0.29649845 0.30306923 0.29667234 0.29544115
## C 0.18866317 0.19702133 0.19519605 0.18773216 0.19395480 0.19369006
## G 0.18863167 0.19715117 0.19528146 0.18775841 0.19417481 0.19380773
## T 0.26346476 0.29292583 0.29673651 0.30316909 0.29739966 0.29532129
## other 0.09616827 0.02053811 0.01628752 0.01827111 0.01779839 0.02173976
## chr7 chr8 chr9 chr10 chr11 chr12
## A 0.28904200 0.29219813 0.2496935 0.28281126 0.28374367 0.28841453
## C 0.19901933 0.19611365 0.1758063 0.20148817 0.20174432 0.19898855
## G 0.19880134 0.19610435 0.1757146 0.20141273 0.20197572 0.19885449
## T 0.28935305 0.29184102 0.2495788 0.28315185 0.28381916 0.28856160
## other 0.02378429 0.02374286 0.1492068 0.03113599 0.02871713 0.02518083
## chr13 chr14 chr15 chr16 chr17 chr18 chr19
## A 0.2547276 0.2421339 0.2303770 0.2404310 0.26060568 0.28773273 0.24337696
## C 0.1598743 0.1679293 0.1682176 0.1951202 0.21833746 0.19005133 0.22794667
## G 0.1598923 0.1683468 0.1680304 0.1959165 0.21799734 0.19037135 0.22850292
## T 0.2554961 0.2440392 0.2301531 0.2415882 0.26118512 0.28804157 0.24402502
## other 0.1700097 0.1775508 0.2032219 0.1269441 0.04187439 0.04380302 0.05614844
## chr20 chr21 chr22 chrX chrY chrM
## A 0.26216449 0.2165582 0.1772703 0.29399618 0.12914207 0.3085511
## C 0.20797651 0.1487685 0.1632600 0.19200905 0.08588285 0.3133184
## G 0.20863631 0.1490699 0.1631285 0.19234703 0.08679432 0.1315551
## T 0.26537230 0.2150178 0.1764863 0.29479139 0.13025126 0.2465753
## other 0.05585039 0.2705855 0.3198550 0.02685635 0.56792951 0.0000000
## chrMT chr4_ctg9_hap1 chr6_apd_hap1 chr6_cox_hap2 chr6_dbb_hap3
## A 3.092522e-01 0.3136227 0.1428621 0.2796939 0.25432414
## C 3.126924e-01 0.1821668 0.1101547 0.2230914 0.20465704
## G 1.309071e-01 0.1815841 0.1104193 0.2237072 0.20502924
## T 2.470879e-01 0.3226264 0.1386412 0.2735075 0.24790734
## other 6.035367e-05 0.0000000 0.4979227 0.0000000 0.08808224
## chr6_mann_hap4 chr6_mcf_hap5 chr6_qbl_hap6 chr6_ssto_hap7 chr17_ctg5_hap1
## A 0.2468544 0.2174387 0.25795168 0.2378417 0.25535867
## C 0.1928190 0.1776351 0.20968048 0.1863057 0.21174623
## G 0.1937628 0.1781635 0.21030038 0.1880758 0.21583589
## T 0.2421801 0.2119062 0.25340743 0.2345850 0.25756472
## other 0.1243838 0.2148565 0.06866004 0.1531918 0.05949449
## chr1_gl000191_random chr1_gl000192_random chr4_gl000193_random
## A 0.2628038 0.2978615 0.2819394
## C 0.2234739 0.2059010 0.2140061
## G 0.2199788 0.2090262 0.2139639
## T 0.2937435 0.2872112 0.2900906
## other 0.0000000 0.0000000 0.0000000
## chr4_gl000194_random chr7_gl000195_random chr8_gl000196_random
## A 0.2765408 0.2923465 0.3557332
## C 0.2168549 0.2024156 0.2032687
## G 0.2157321 0.2042089 0.1932209
## T 0.2908722 0.3010290 0.2477771
## other 0.0000000 0.0000000 0.0000000
## chr8_gl000197_random chr9_gl000198_random chr9_gl000199_random
## A 0.23252186 0.3090748 0.3220151
## C 0.26585071 0.1733585 0.2059232
## G 0.27276395 0.2051951 0.1732225
## T 0.22617350 0.3123716 0.2988391
## other 0.00268998 0.0000000 0.0000000
## chr9_gl000200_random chr9_gl000201_random chr11_gl000202_random
## A 0.2959500 0.1964424 0.2300576
## C 0.1989039 0.2869592 0.2806274
## G 0.2005721 0.3074582 0.2654415
## T 0.3045740 0.2091402 0.2238735
## other 0.0000000 0.0000000 0.0000000
## chr17_gl000203_random chr17_gl000204_random chr17_gl000205_random
## A 0.3350579 0.2423072 0.2821213
## C 0.1619820 0.2791908 0.2103008
## G 0.1708891 0.2654655 0.2070360
## T 0.3320710 0.2130365 0.3005418
## other 0.0000000 0.0000000 0.0000000
## chr17_gl000206_random chr18_gl000207_random chr19_gl000208_random
## A 0.2138972 0.2355702 0.3148054
## C 0.2556767 0.1379634 0.1981789
## G 0.2752372 0.3015016 0.1784354
## T 0.2551889 0.3249648 0.3085803
## other 0.0000000 0.0000000 0.0000000
## chr19_gl000209_random chr21_gl000210_random chrUn_gl000211 chrUn_gl000212
## A 0.2762975 0.227151217 0.3057407 0.2821875
## C 0.2226753 0.279098331 0.1919239 0.2271993
## G 0.2422896 0.261939166 0.1951599 0.2166458
## T 0.2587376 0.228198830 0.3071755 0.2739674
## other 0.0000000 0.003612456 0.0000000 0.0000000
## chrUn_gl000213 chrUn_gl000214 chrUn_gl000215 chrUn_gl000216
## A 0.2925432 0.2951321 0.2917152 0.2403392
## C 0.2059864 0.1995672 0.2100901 0.2711470
## G 0.2030334 0.2156436 0.2099336 0.1485948
## T 0.2984370 0.2896571 0.2882610 0.3399190
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000217 chrUn_gl000218 chrUn_gl000219 chrUn_gl000220
## A 0.3017444 0.2856398 0.3043003 0.2300960
## C 0.1892430 0.2066188 0.1981105 0.2516656
## G 0.1866465 0.2095850 0.2014978 0.2329823
## T 0.3223661 0.2981563 0.2960915 0.2852561
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000221 chrUn_gl000222 chrUn_gl000223 chrUn_gl000224
## A 0.3083393 0.2757825 0.2933197 0.2796325
## C 0.1923203 0.2186973 0.2152836 0.2082997
## G 0.1940321 0.2210413 0.2167133 0.2245775
## T 0.3053083 0.2844788 0.2746834 0.2874903
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000225 chrUn_gl000226 chrUn_gl000227 chrUn_gl000228
## A 0.2696509 0.2999733 0.3199713 0.2363073
## C 0.2317105 0.1749733 0.2044807 0.2748993
## G 0.2448230 0.2152852 0.2056180 0.2644517
## T 0.2538156 0.3097681 0.2699300 0.2243417
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000229 chrUn_gl000230 chrUn_gl000231 chrUn_gl000232
## A 0.2000201 0.2901742 0.2592566 0.2837253
## C 0.2704264 0.2137282 0.2265026 0.2088458
## G 0.2310551 0.2033828 0.2201490 0.2093870
## T 0.2984985 0.2927147 0.2940919 0.2980419
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000233 chrUn_gl000234 chrUn_gl000235 chrUn_gl000236
## A 0.2653839 0.2663887 0.3435923 0.3101541
## C 0.2068740 0.2152673 0.1910135 0.2010779
## G 0.2173440 0.2154400 0.1889540 0.2152430
## T 0.3103981 0.3029040 0.2764402 0.2735251
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000237 chrUn_gl000238 chrUn_gl000239 chrUn_gl000240
## A 0.2675780 0.2604973 0.2726171 0.3243508
## C 0.2232760 0.1954230 0.2148770 0.2137219
## G 0.2433558 0.2045870 0.2391497 0.2117664
## T 0.2657902 0.3394927 0.2733562 0.2501610
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000241 chrUn_gl000242 chrUn_gl000243 chrUn_gl000244
## A 0.3231163 0.2433426 0.2475254 0.2741867
## C 0.1865629 0.2305218 0.2377426 0.2144807
## G 0.1863020 0.2534292 0.2223299 0.2218187
## T 0.3040188 0.2727064 0.2924021 0.2895139
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000245 chrUn_gl000246 chrUn_gl000247 chrUn_gl000248
## A 0.3512865 0.2736279 0.3020702 0.2542100
## C 0.1829964 0.1927452 0.2139915 0.2328709
## G 0.1801315 0.1937412 0.2220087 0.2236968
## T 0.2855857 0.3398857 0.2619296 0.2892223
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrUn_gl000249 chr1_gl383516_fix chr1_gl383517_fix chr1_gl949741_fix
## A 0.2803231 0.3008963 0.3051345 0.2516645
## C 0.2331827 0.2175764 0.2172759 0.2265574
## G 0.2346112 0.2133385 0.2081982 0.2368312
## T 0.2518830 0.2681888 0.2693913 0.2849470
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr1_jh636052_fix chr1_jh636053_fix chr1_jh636054_fix chr1_jh806573_fix
## A 0.2915854 0.2974645 0.2677319 0.2461507
## C 0.2111091 0.2018625 0.2188790 0.2453809
## G 0.2092759 0.2037353 0.2276569 0.2471232
## T 0.2880296 0.2969377 0.2857322 0.2613452
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr1_jh806574_fix chr1_jh806575_fix chr2_gl877870_fix chr2_gl877871_fix
## A 0.2163867 0.2903457 0.2862271 0.3017369
## C 0.2863545 0.2308422 0.1949380 0.2067067
## G 0.2504569 0.2398912 0.2080702 0.1945740
## T 0.2468018 0.2389209 0.3107648 0.2969823
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr2_kb663603_fix chr3_gl383523_fix chr3_gl383524_fix chr3_gl383525_fix
## A 0.2805614 0.2872107 0.3484320 0.2938536
## C 0.2292188 0.2327587 0.1856637 0.1877104
## G 0.2218436 0.2240462 0.1854606 0.2034643
## T 0.2683762 0.2559844 0.2804437 0.3149716
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr3_jh159131_fix chr3_jh159132_fix chr3_ke332495_fix chr4_gl582967_fix
## A 0.3129855 0.2165372 0.2651169 0.3151178
## C 0.1983980 0.2801160 0.2341839 0.1859600
## G 0.1874119 0.2894214 0.2359235 0.1836431
## T 0.3012045 0.2139254 0.2647758 0.3152790
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr4_gl877872_fix chr4_ke332496_fix chr5_jh159133_fix chr5_ke332497_fix
## A 0.2260887 0.2827837 0.3082428 0.2927088
## C 0.1943527 0.2041732 0.1755734 0.1969190
## G 0.1875389 0.2058345 0.1859032 0.2026945
## T 0.2239441 0.3072086 0.3302806 0.3076777
## other 0.1680757 0.0000000 0.0000000 0.0000000
## chr6_jh636056_fix chr6_jh636057_fix chr6_jh806576_fix chr6_kb663604_fix
## A 0.3020745 0.2886735 0.3036915 0.2848935
## C 0.1926120 0.2259797 0.1991543 0.2214604
## G 0.2033532 0.2242963 0.2004528 0.2207861
## T 0.3019604 0.2610505 0.2967014 0.2728599
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr6_ke332498_fix chr7_gl582968_fix chr7_gl582969_fix chr7_gl582970_fix
## A 0.2705915 0.2479387 0.2768214 0.2658675
## C 0.2335874 0.2494963 0.2144562 0.2369186
## G 0.2294320 0.2438807 0.2261152 0.2417782
## T 0.2663892 0.2586844 0.2826072 0.2554357
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr7_gl582971_fix chr7_gl582972_fix chr7_jh159134_fix chr7_jh636058_fix
## A 0.2906429 0.2567043 0.2583319 0.27316619
## C 0.2084313 0.2369437 0.2433786 0.19363274
## G 0.2091196 0.2416360 0.2418288 0.19167666
## T 0.2918062 0.2647159 0.2564607 0.27171414
## other 0.0000000 0.0000000 0.0000000 0.06981027
## chr7_ke332499_fix chr8_gl383535_fix chr8_gl383536_fix chr8_gl949743_fix
## A 0.2797819 0.2138011 0.2026136 0.2739874
## C 0.2016203 0.2945841 0.2998523 0.2159851
## G 0.2079076 0.2775717 0.2945769 0.2216656
## T 0.3106903 0.2140431 0.2029572 0.2883619
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr8_jh159135_fix chr8_ke332500_fix chr9_gl339450_fix chr9_gl383537_fix
## A 0.2633324 0.2809599 0.2434184 0.2157284
## C 0.2508435 0.1932923 0.2550975 0.3011132
## G 0.2468827 0.2071417 0.2654469 0.2955073
## T 0.2389414 0.3186061 0.2360372 0.1876512
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr9_gl383538_fix chr9_jh636059_fix chr9_jh806577_fix chr9_jh806578_fix
## A 0.1844524 0.2905386 0.1904528 0.2116067
## C 0.3013535 0.1947803 0.2885148 0.2722074
## G 0.3299040 0.2047844 0.2922658 0.2924155
## T 0.1842901 0.3098968 0.2287666 0.2237705
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr9_jh806579_fix chr9_kb663605_fix chr10_gl383543_fix chr10_gl383544_fix
## A 0.2186676 0.2543835 0.2838067 0.2633940
## C 0.2610420 0.2338289 0.1968497 0.2307327
## G 0.2660016 0.2450457 0.2024736 0.2340043
## T 0.2542888 0.2667419 0.3168700 0.2718690
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr10_gl877873_fix chr10_jh591181_fix chr10_jh591182_fix
## A 0.2985961 0.2759234 0.2662309
## C 0.2143650 0.2127839 0.2418909
## G 0.2068798 0.2125819 0.2326125
## T 0.2801591 0.2767918 0.2592657
## other 0.0000000 0.0219190 0.0000000
## chr10_jh591183_fix chr10_jh636060_fix chr10_jh806580_fix
## A 0.3060196 0.3087915 0.2414197
## C 0.2057104 0.2039293 0.2332822
## G 0.1943402 0.2009312 0.2601209
## T 0.2939299 0.2863481 0.2651773
## other 0.0000000 0.0000000 0.0000000
## chr10_kb663606_fix chr10_ke332501_fix chr11_gl582973_fix
## A 0.2986107 0.2941566 0.3148621
## C 0.2044067 0.1797307 0.1998044
## G 0.2050768 0.1852263 0.1994710
## T 0.2919059 0.3408864 0.2858625
## other 0.0000000 0.0000000 0.0000000
## chr11_gl949744_fix chr11_jh159138_fix chr11_jh159139_fix
## A 0.2458111 0.2491665 0.3059755
## C 0.2546519 0.2476326 0.1944936
## G 0.2557805 0.2495982 0.2053786
## T 0.2437565 0.2536028 0.2941523
## other 0.0000000 0.0000000 0.0000000
## chr11_jh159140_fix chr11_jh159141_fix chr11_jh159142_fix
## A 0.2820317 0.2886761 0.2932768
## C 0.2082956 0.2019977 0.2117332
## G 0.2137015 0.2081570 0.2094432
## T 0.2959712 0.3011691 0.2855468
## other 0.0000000 0.0000000 0.0000000
## chr11_jh159143_fix chr11_jh591184_fix chr11_jh591185_fix
## A 0.2365075 0.2923194 0.2985959
## C 0.2503579 0.2114402 0.2000872
## G 0.2598405 0.2046478 0.1953989
## T 0.2532941 0.2915926 0.3059180
## other 0.0000000 0.0000000 0.0000000
## chr11_jh720443_fix chr11_jh806581_fix chr12_gl383548_fix
## A 0.2679578 0.24112875 0.2291055
## C 0.2261807 0.24222035 0.3036061
## G 0.2262468 0.23897651 0.2740564
## T 0.2796146 0.22034250 0.1932319
## other 0.0000000 0.05733189 0.0000000
## chr12_gl582974_fix chr12_jh720444_fix chr12_kb663607_fix
## A 0.2463166 2.418134e-01 0.2652498
## C 0.2548102 2.476934e-01 0.2465738
## G 0.2572781 2.525153e-01 0.2403784
## T 0.2415951 2.579706e-01 0.2477980
## other 0.0000000 7.322574e-06 0.0000000
## chr13_gl582975_fix chr14_kb021645_fix chr15_jh720445_fix
## A 0.2865963 0.2550673 0.2652367
## C 0.2060181 0.2350002 0.2363247
## G 0.2179620 0.2376574 0.2272500
## T 0.2894236 0.2722751 0.2711885
## other 0.0000000 0.0000000 0.0000000
## chr16_jh720446_fix chr17_gl383558_fix chr17_gl383559_fix
## A 0.2416046 0.2936936 0.2448234
## C 0.2709230 0.2213674 0.2446551
## G 0.2624788 0.2134601 0.2519755
## T 0.2249936 0.2714789 0.2585459
## other 0.0000000 0.0000000 0.0000000
## chr17_gl383560_fix chr17_gl383561_fix chr17_gl383562_fix
## A 0.2546136 0.28450789 0.2663827
## C 0.2327602 0.19281220 0.2400825
## G 0.2375835 0.24846026 0.2448025
## T 0.2750427 0.24481980 0.2487322
## other 0.0000000 0.02939985 0.0000000
## chr17_gl582976_fix chr17_jh159144_fix chr17_jh159145_fix
## A 0.2504660 0.2686409 0.2546161
## C 0.2630225 0.2237369 0.2467233
## G 0.2519859 0.2279085 0.2446706
## T 0.2345256 0.2797137 0.2539900
## other 0.0000000 0.0000000 0.0000000
## chr17_jh591186_fix chr17_jh636061_fix chr17_jh720447_fix
## A 0.2507981 0.1990981 0.2407870
## C 0.2722189 0.2965403 0.1963929
## G 0.2609596 0.3012109 0.2016088
## T 0.2160235 0.2031506 0.2511725
## other 0.0000000 0.0000000 0.1100388
## chr17_jh806582_fix chr17_kb021646_fix chr17_ke332502_fix
## A 0.2539408 0.2601411 0.32603479
## C 0.2662980 0.2252431 0.15536183
## G 0.2486786 0.2397974 0.23171852
## T 0.2310826 0.2748184 0.27681498
## other 0.0000000 0.0000000 0.01006988
## chr19_gl582977_fix chr19_jh159149_fix chr19_kb021647_fix
## A 0.2977965 0.2410897 0.2550652
## C 0.2001058 0.2400386 0.2425837
## G 0.2048560 0.2433099 0.2469098
## T 0.2972417 0.2755619 0.2554412
## other 0.0000000 0.0000000 0.0000000
## chr19_ke332505_fix chr20_gl582979_fix chr20_jh720448_fix
## A 0.2664916 0.1967882 0.220989458
## C 0.2467141 0.2930533 0.252912050
## G 0.2376043 0.2975114 0.268589589
## T 0.2491900 0.2126471 0.252571542
## other 0.0000000 0.0000000 0.004937361
## chr20_kb663608_fix chr21_ke332506_fix chr22_jh720449_fix
## A 0.3067808 0.2671325 0.2765028
## C 0.1854622 0.2528869 0.2395972
## G 0.1888408 0.2468235 0.2310432
## T 0.3189162 0.2331571 0.2528568
## other 0.0000000 0.0000000 0.0000000
## chr22_jh806583_fix chr22_jh806584_fix chr22_jh806585_fix
## A 0.3140870 0.2865427 0.3446296
## C 0.1935843 0.2174643 0.1635127
## G 0.1818666 0.2063181 0.2360112
## T 0.3104622 0.2896749 0.2558465
## other 0.0000000 0.0000000 0.0000000
## chr22_jh806586_fix chrX_gl877877_fix chrX_jh159150_fix chrX_jh720451_fix
## A 0.2662655 0.2681925 0.2668093 0.3067891
## C 0.2189330 0.2297814 0.2359276 0.1897542
## G 0.2372138 0.2284001 0.2352818 0.1841511
## T 0.2775877 0.2736261 0.2619812 0.3193056
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrX_jh720452_fix chrX_jh720453_fix chrX_jh720454_fix chrX_jh720455_fix
## A 0.2968263 0.3135900 0.3163970 0.3033951
## C 0.1945880 0.1965462 0.1990463 0.1957745
## G 0.1960832 0.1929587 0.1946928 0.1976351
## T 0.3125025 0.2969050 0.2898638 0.3031953
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrX_jh806587_fix chrX_jh806588_fix chrX_jh806589_fix chrX_jh806590_fix
## A 0.2942685 0.3114334 0.3090234 0.2586284
## C 0.2089787 0.1854448 0.2036507 0.2109463
## G 0.2081990 0.1840315 0.1996342 0.2131978
## T 0.2885538 0.3190903 0.2876917 0.2634729
## other 0.0000000 0.0000000 0.0000000 0.0537547
## chrX_jh806591_fix chrX_jh806592_fix chrX_jh806593_fix chrX_jh806594_fix
## A 0.3195958 0.2987292 0.2897511 0.2874012
## C 0.1971436 0.2058772 0.2028997 0.2075181
## G 0.1928549 0.2015920 0.2090054 0.2064477
## T 0.2904058 0.2938016 0.2983438 0.2986330
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrX_jh806595_fix chrX_jh806596_fix chrX_jh806597_fix chrX_jh806598_fix
## A 0.2834190 0.2896332 0.2917909 0.3067128
## C 0.2088728 0.2112015 0.2074660 0.1912189
## G 0.2105865 0.2112208 0.2095901 0.1945692
## T 0.2971217 0.2879445 0.2911530 0.3074990
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrX_jh806599_fix chrX_jh806600_fix chrX_jh806601_fix chrX_jh806602_fix
## A 0.3180280 0.3058339 0.3191182 0.2962429
## C 0.1846529 0.1961708 0.1871598 0.1952624
## G 0.1845574 0.1967587 0.1865245 0.1969448
## T 0.3127617 0.3012365 0.3071975 0.3115500
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chrX_jh806603_fix chrX_kb021648_fix chr1_gl383518_alt chr1_gl383519_alt
## A 0.3150113 0.2737418 0.2608598 0.2446766
## C 0.1808646 0.2168874 0.2366380 0.2563844
## G 0.1878556 0.2170427 0.2339357 0.2585700
## T 0.3162685 0.2923281 0.2685665 0.2403689
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr1_gl383520_alt chr2_gl383521_alt chr2_gl383522_alt chr2_gl582966_alt
## A 0.3039099 0.2988214 0.3154069 0.2649926
## C 0.1779453 0.1902015 0.2017348 0.2021200
## G 0.1801167 0.1989190 0.1917042 0.2286151
## T 0.3380281 0.3120580 0.2911542 0.3042723
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr3_gl383526_alt chr3_jh636055_alt chr4_gl383527_alt chr4_gl383528_alt
## A 0.3027547 0.2893371 0.3128252 0.3349345
## C 0.1840140 0.2191671 0.1860018 0.1736317
## G 0.1894715 0.2162852 0.1856797 0.1688894
## T 0.3237598 0.2752107 0.3154933 0.3225444
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr4_gl383529_alt chr5_gl339449_alt chr5_gl383530_alt chr5_gl383531_alt
## A 0.3199473 0.28141802 0.2974190 0.3242726
## C 0.1555812 0.18531453 0.1816853 0.1813051
## G 0.2027607 0.18635240 0.1909503 0.1847929
## T 0.3217108 0.28491600 0.3299454 0.3096294
## other 0.0000000 0.06199905 0.0000000 0.0000000
## chr5_gl383532_alt chr5_gl949742_alt chr6_gl383533_alt chr6_kb021644_alt
## A 0.3265158 0.3179298 0.3108886 0.2491961
## C 0.1739314 0.1752200 0.1928313 0.2111658
## G 0.1697249 0.1729718 0.1924384 0.2723933
## T 0.3298279 0.3338785 0.3038417 0.2672449
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr7_gl383534_alt chr9_gl383539_alt chr9_gl383540_alt chr9_gl383541_alt
## A 0.2928270 0.2920767 0.2582913 0.2846292
## C 0.2071772 0.1931615 0.2262582 0.2024976
## G 0.2059774 0.2033094 0.2304091 0.2068762
## T 0.2940184 0.3114524 0.2850414 0.3059970
## other 0.0000000 0.0000000 0.0000000 0.0000000
## chr9_gl383542_alt chr10_gl383545_alt chr10_gl383546_alt
## A 0.3008229 0.3003559 0.3054080
## C 0.2096049 0.1990862 0.2153279
## G 0.2174674 0.1978756 0.2112672
## T 0.2721049 0.3026822 0.2679970
## other 0.0000000 0.0000000 0.0000000
## chr11_gl383547_alt chr11_jh159136_alt chr11_jh159137_alt
## A 0.3173820 0.3304759 0.3196036
## C 0.1701607 0.1770764 0.1883767
## G 0.1712293 0.1754097 0.1842599
## T 0.3412281 0.3170380 0.3077598
## other 0.0000000 0.0000000 0.0000000
## chr12_gl383549_alt chr12_gl383550_alt chr12_gl383551_alt
## A 0.3163885 0.3125879 0.2818809
## C 0.1952005 0.2043469 0.1980805
## G 0.1909705 0.1912424 0.2071463
## T 0.2974405 0.2918228 0.3128923
## other 0.0000000 0.0000000 0.0000000
## chr12_gl383552_alt chr12_gl383553_alt chr12_gl877875_alt
## A 0.3060762 0.2847051 0.2497475
## C 0.1797916 0.1940160 0.2524370
## G 0.1853882 0.1997397 0.2547082
## T 0.3287440 0.3215393 0.2431072
## other 0.0000000 0.0000000 0.0000000
## chr12_gl877876_alt chr12_gl949745_alt chr15_gl383554_alt
## A 0.3264866 0.3284864 0.2816135
## C 0.1866799 0.1863133 0.2170426
## G 0.1764539 0.1769388 0.2162029
## T 0.3103796 0.3082615 0.2851410
## other 0.0000000 0.0000000 0.0000000
## chr15_gl383555_alt chr16_gl383556_alt chr16_gl383557_alt
## A 0.2660010 0.2907743 0.3029374
## C 0.2473989 0.2263668 0.2221541
## G 0.2416114 0.2177936 0.2162548
## T 0.2449887 0.2650653 0.2586538
## other 0.0000000 0.0000000 0.0000000
## chr17_gl383563_alt chr17_gl383564_alt chr17_gl383565_alt
## A 0.2359719 0.2637307 0.3179000
## C 0.2819238 0.2279818 0.1853702
## G 0.2535179 0.2273359 0.1858925
## T 0.2285864 0.2809517 0.3108373
## other 0.0000000 0.0000000 0.0000000
## chr17_gl383566_alt chr17_jh159146_alt chr17_jh159147_alt
## A 0.2286215 0.2889214 0.2720449
## C 0.2579723 0.1956596 0.2029711
## G 0.2632594 0.2072189 0.2047338
## T 0.2501469 0.3082001 0.3202502
## other 0.0000000 0.0000000 0.0000000
## chr17_jh159148_alt chr18_gl383567_alt chr18_gl383568_alt
## A 0.2740547 0.2771063 0.3189418
## C 0.1994663 0.2079281 0.1905846
## G 0.2059271 0.2091219 0.1871222
## T 0.3205518 0.3058438 0.3033514
## other 0.0000000 0.0000000 0.0000000
## chr18_gl383569_alt chr18_gl383570_alt chr18_gl383571_alt
## A 0.2778208 0.3148693 0.2407226
## C 0.2426139 0.1954499 0.1334944
## G 0.2356237 0.1955652 0.1375997
## T 0.2439416 0.2941155 0.2360121
## other 0.0000000 0.0000000 0.2521712
## chr18_gl383572_alt chr19_gl383573_alt chr19_gl383574_alt
## A 0.2603433 0.2983221 0.2688241
## C 0.2419162 0.2124634 0.2153159
## G 0.2300639 0.1924767 0.2229764
## T 0.2676766 0.2967378 0.2928835
## other 0.0000000 0.0000000 0.0000000
## chr19_gl383575_alt chr19_gl383576_alt chr19_gl949746_alt
## A 0.3049782 0.2951485 0.2260468
## C 0.2015016 0.1956931 0.2190468
## G 0.2015368 0.1992618 0.2218462
## T 0.2919834 0.3098966 0.2267545
## other 0.0000000 0.0000000 0.1063059
## chr19_gl949747_alt chr19_gl949748_alt chr19_gl949749_alt
## A 0.22975413 0.1063870 0.09904748
## C 0.22236432 0.1068483 0.09982232
## G 0.22754719 0.1099687 0.10185558
## T 0.22986379 0.1064697 0.09845399
## other 0.09047057 0.5703263 0.60082063
## chr19_gl949750_alt chr19_gl949751_alt chr19_gl949752_alt
## A 0.09667579 0.1156837 0.253165839
## C 0.09859723 0.1157366 0.241468949
## G 0.09985756 0.1184882 0.246083477
## T 0.09721124 0.1155252 0.254216391
## other 0.60765818 0.5345663 0.005065343
## chr19_gl949753_alt chr20_gl383577_alt chr21_gl383578_alt
## A 0.2145533 0.2764186 0.3033622
## C 0.2090529 0.2404175 0.1791229
## G 0.2116053 0.2383690 0.1825180
## T 0.2141252 0.2447950 0.3349969
## other 0.1506633 0.0000000 0.0000000
## chr21_gl383579_alt chr21_gl383580_alt chr21_gl383581_alt
## A 0.3180002 0.2780502 0.2941126
## C 0.1705087 0.2086883 0.2192733
## G 0.1782672 0.2078176 0.2259748
## T 0.3332240 0.3054439 0.2606393
## other 0.0000000 0.0000000 0.0000000
## chr22_gl383582_alt chr22_gl383583_alt chr22_kb663609_alt
## A 0.2722482 0.2601007 0.2319998
## C 0.2369987 0.2331621 0.2528610
## G 0.2315630 0.2279208 0.2526448
## T 0.2591901 0.2788164 0.2624944
## other 0.0000000 0.0000000 0.0000000Biostrings
- Here are two ways to ask if a specific pattern is present in one sequence \({\tt matchPattern}\) or a set of sequences \({\tt vmatchPattern}\). Pattern matching and alignments are very central in biology.
chr2 <- Hsapiens$chr2
matchPattern( "ATGATAAAAGAA", chr2 )
## Views on a 243199373-letter DNAString subject
## subject: NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN...NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
## views:
## start end width
## [1] 3093768 3093779 12 [ATGATAAAAGAA]
## [2] 3924476 3924487 12 [ATGATAAAAGAA]
## [3] 4662559 4662570 12 [ATGATAAAAGAA]
## [4] 4967998 4968009 12 [ATGATAAAAGAA]
## [5] 7734903 7734914 12 [ATGATAAAAGAA]
## ... ... ... ... ...
## [85] 218329792 218329803 12 [ATGATAAAAGAA]
## [86] 227061871 227061882 12 [ATGATAAAAGAA]
## [87] 233071842 233071853 12 [ATGATAAAAGAA]
## [88] 240711797 240711808 12 [ATGATAAAAGAA]
## [89] 240760809 240760820 12 [ATGATAAAAGAA]
vmatchPattern( "ATGATAAAAGAA", Hsapiens)
## GRanges object with 2313 ranges and 0 metadata columns:
## seqnames ranges strand
## <Rle> <IRanges> <Rle>
## [1] chr1 4721174-4721185 +
## [2] chr1 17504484-17504495 +
## [3] chr1 20292323-20292334 +
## [4] chr1 21316299-21316310 +
## [5] chr1 25001061-25001072 +
## ... ... ... ...
## [2309] chr9_gl383541_alt 90799-90810 +
## [2310] chr11_jh159136_alt 61724-61735 +
## [2311] chr11_jh159136_alt 92679-92690 -
## [2312] chr11_jh159136_alt 199693-199704 -
## [2313] chr12_gl383550_alt 144547-144558 +
## -------
## seqinfo: 298 sequences from an unspecified genomeBiostrings
- This is also an example of an alignment, but here N matches any nucleotide. This is an example of a regular expression.
PpiI <- "GAACNNNNNCTC" # a restriction enzyme pattern
(align.PpiI <- matchPattern(PpiI, Hsapiens[[1]][17000:23000], fixed=FALSE))
## Views on a 6001-letter DNAString subject
## subject: ACATCAATCTCAGGCACCTGGCCCAGGTCTGGCA...CTTAAAGACCTGCATCCTCTTCCCTAGGTGTCCC
## views:
## start end width
## [1] 5695 5706 12 [GAACCCACACTC]Biostrings
- There are more biologically relevant notions of alignment that ultimately are based in models of evolution. We will learn these in part 3 of the course.
Biostrings
We will later explore several different notions of alignment.
BLAST is perhaps the best known “industrial strength” tool for alignments (available at the NCBI).
Annotations
We can annotate the genomes. For example, we can include single nucleotide polymorphims (mutations that have been observed between indivduals).
Large-scale GWAS reveals insights into the genetic architecture of same-sex sexual behavior
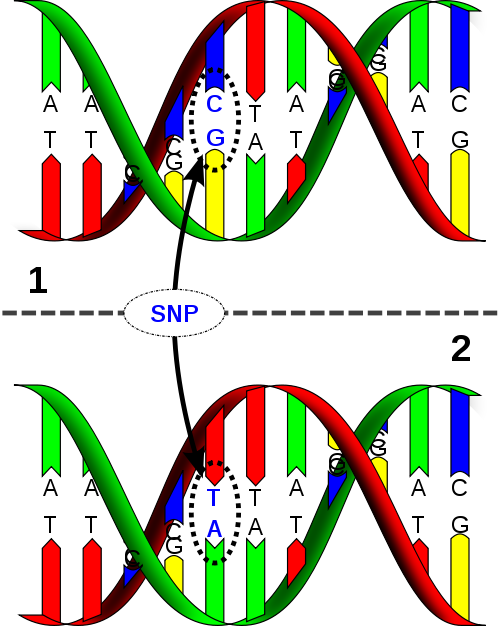
Annotations
available.SNPs()
## [1] "SNPlocs.Hsapiens.dbSNP.20101109"
## [2] "SNPlocs.Hsapiens.dbSNP.20120608"
## [3] "SNPlocs.Hsapiens.dbSNP141.GRCh38"
## [4] "SNPlocs.Hsapiens.dbSNP142.GRCh37"
## [5] "SNPlocs.Hsapiens.dbSNP144.GRCh37"
## [6] "SNPlocs.Hsapiens.dbSNP144.GRCh38"
## [7] "SNPlocs.Hsapiens.dbSNP149.GRCh38"
## [8] "SNPlocs.Hsapiens.dbSNP150.GRCh38"
## [9] "SNPlocs.Hsapiens.dbSNP151.GRCh38"
## [10] "XtraSNPlocs.Hsapiens.dbSNP141.GRCh38"
## [11] "XtraSNPlocs.Hsapiens.dbSNP144.GRCh37"
## [12] "XtraSNPlocs.Hsapiens.dbSNP144.GRCh38"
# BiocManager::install("SNPlocs.Hsapiens.dbSNP.20101109")
library(SNPlocs.Hsapiens.dbSNP.20101109)
## Please note that the SNPlocs.Hsapiens.dbSNP.20101109 package contains
## outdated dbSNP data and will be deprecated in the near future. We
## highly recommend that you use a SNPlocs package based on a more recent
## dbSNP build for your analyses instead. See available.SNPs() for the
## list of SNPlocs packages currently available and make sure to pick up
## the most recent one.
SnpHsapiens <- injectSNPs(Hsapiens, "SNPlocs.Hsapiens.dbSNP.20101109")
snpcount(SnpHsapiens)
## chr1 chr2 chr3 chr4 chr5 chr6 chr7 chr8 chr9 chr10
## 1849438 1936836 1613418 1613633 1453710 1446827 1335745 1243129 995075 1158707
## chr11 chr12 chr13 chr14 chr15 chr16 chr17 chr18 chr19 chr20
## 1147722 1105364 815729 740129 657719 757926 641905 645646 520666 586708
## chr21 chr22 chrX chrY
## 338254 331060 529608 67438
head(snplocs(SnpHsapiens, "chr1"))
## RefSNP_id alleles_as_ambig loc
## 1 112750067 Y 10327
## 2 112155239 M 10440
## 3 117577454 S 10469
## 4 55998931 Y 10492
## 5 62636508 S 10519
## 6 114315702 S 10533Addition packages
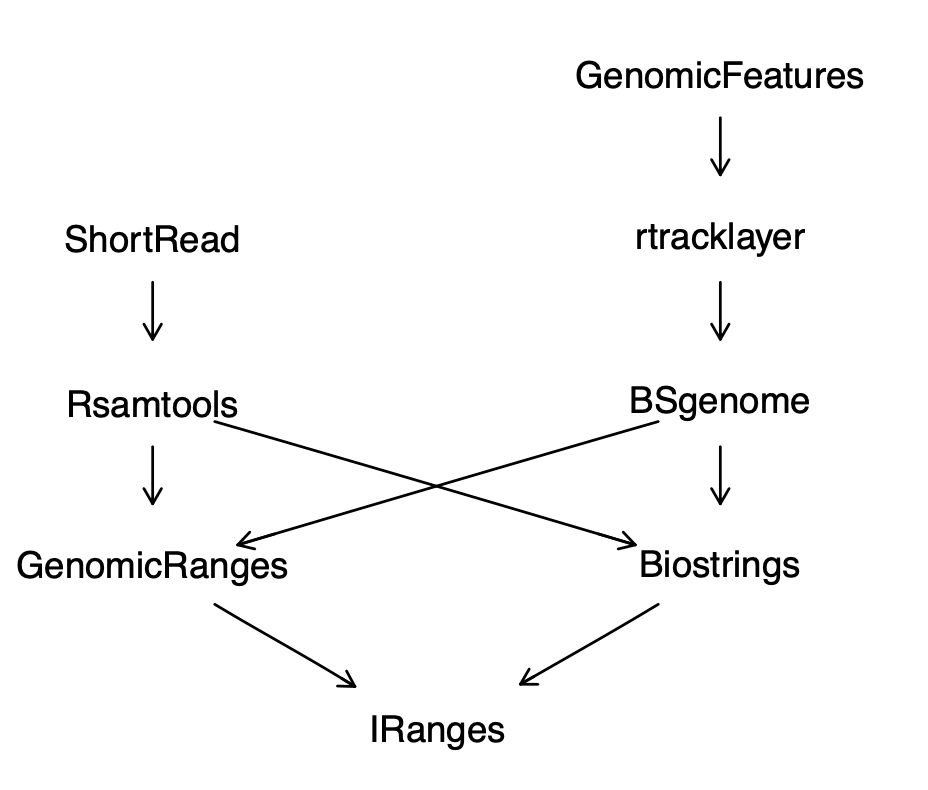
There are several additional packages that are very important for deadling with large genomes, especially human.
\({\tt annotatR}\) is great for epigenetics and SNP (single nucleotide polymorphism) studies.
\({\tt rtracklayer}\) can create and plot tracks like we see in the UCSC genome browser.
Genome and annotations for S. cerevisiae
Baker’s yeast has served as a very important model organism.
The genomes of this small fungi (eukaryote) has 16 chromosomes and one mitochondrial genome (total \(12.1M\) bp). In total \(6,275\) genes.

S. cerevisiae
SGD has long served as an excellent curated bioinformatics resource.
Let’s go live to RStudio Cloud and download its genome and annotations, and wrangle them into R.
Points of reflection
Why are roman numerals not a great idea?
Why is it perhaps inconvenient to have both a portion of a file in \({\tt gff}\) format and a portion in \({\tt FASTA}\) format? What sort of punishment would be appropriate for this type of design decision?
What is the difference between a gene, transcript and CDS at SGD?
What are some of the different types of entities that are cataloged and mapped to the genome of baker’s yeast?
BIOL 480
© M Hallett, 2020 Concordia University
